Resumen
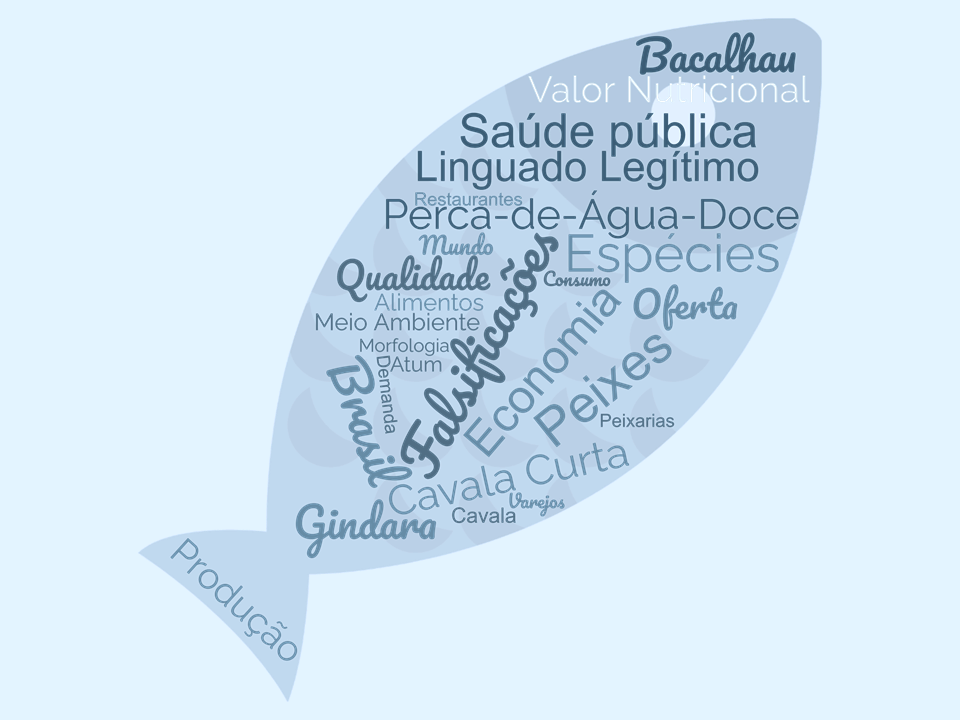
O objetivo deste artigo é determinar quais são as espécies de peixes mais comumente substituídas, relatadas em revisões sobre falsificações por troca de espécies, em artigos científicos publicados entre 2002 e 2022, assim como expor quais são os fatores que levam à ocorrência de casos de falsificação, quais os danos sociais que tal prática ilegal pode gerar e em que tipo de estabelecimentos as falsificações normalmente são detectadas. Assim, por meio de uma revisão sistemática de literatura, foi verificado que as espécies de peixes mais comumente falsificadas foram Anoplopoma fimbria, Gadus morhua, Solea solea, Thunnus albacares, Scomberomorus commerson, Lates calcarifer e Rastrelliger brachysoma. As motivações por trás das falsificações intencionais foram predominantemente econômicas, tendo sido observados impactos negativos das falsificações na economia, na saúde pública e no meio ambiente. A maioria das falsificações foi encontrada no final da cadeia produtiva, em locais como varejos, restaurantes e peixarias. Dessa forma, ao saber como as falsificações acontecem e suas razões, torna-se possível saber como minimizar a sua ocorrência.
Citas
1. Sartori AGO, Amancio RD. Pescado: importância nutricional e consumo no Brasil. Seg Alim Nutr. 2012;19(2):83-93.https://doi.org/10.20396/san.v19i2.8634613
2. Harvard TH Chan, School of Public Health. Omega-3 fatty acids: an essential contribution. Boston (MA): University of Harvard, School of Public Health; 2023. Disponível em: https://www.hsph.harvard.edu/nutritionsource/what-should-you-eat/fats-and-cholesterol/types-of-fat/omega-3-fats/#:~:text=The%20human%20body%20can%20make,must%20get%20them%20from%20food
3. Carrera E, Terni M, Montero A, Garcia T, Gonzalez I, Martin R. ELISA-based detection of mislabeled albacore (Thunnus alalunga) fresh and frozen fish fillets. Food Agric Immunol. 2014;25(4):569-77. https://doi.org/10.1080/09540105.2013.858310
4. Cawthorn DM, Duncan J, Kastern C, Francis J, Hoffman LC. Fish species substitution and misnaming in South Africa: an economic, safety and sustainability conundrum revisited. Food Chem.2015;185:165-81. https://doi.org/10.1016/j.foodchem.2015.03.113
5. Atos do Poder Executivo (BR). Decreto nº 10.468, de 18 de agosto de 2020. Altera o Decreto nº 9.013, de 29 de março de 2017, que regulamenta a Lei nº 1.283, de 18 de dezembro de 1950, e a Lei nº 7.889, de 23 de novembro de 1989, que dispõem sobre o regulamento da inspeção industrial e sanitária de produtos de origem animal. Diário Oficial da União. Brasília, DF, 19 ago 2020. Seção 1(159):5-14.
6. Bénard-Capelle J, Guillonneau V, Nouvian C, Fournier N, Le Loët K, Dettai A. Fish mislabelling in France: substitution rates and retail types. Peer J. 2015;2:e714. https://doi.org/10.7717/peerj.714
7. Horreo JL, Fitze PS, Jiménez-Valverde A, Noriega JA, Pelaez ML. Amplification of 16S rDNA reveals important fish mislabeling in Madrid restaurants. Food Control. 2019;96:146-50. https://doi.org/10.1016/j.foodcont.2018.09.020
8. Munguia-Vega A, Weaver AH, Dominguez-Contreras JF, Peckham H. Multiple drivers behind mislabeling of fish from artisanal fisheries in La Paz, Mexico. Peer J. 2021;9:e10750. https://doi.org/10.7717/peerj.10750
9. Acutis PL, Cambiotti V, Riina MV, Meistro S, Maurella C, Massaro M et al. Detection of fish species substitution frauds in Italy: a targeted national monitoring plan. Food Control. 2019;101:151-5. https://doi.org/10.1016/j.foodcont.2019.02.020
10. Blanco-Fernandez C, Ardura A, Masiá P, Rodriguez N, Voces L, Fernandez-Raigoso M et al. Fraud in highly appreciated fish detected from DNA in Europe may undermine the development goal of sustainable fishing in Africa. Sci Rep. 2021;11(1):11423. https://doi.org/10.1038/s41598-021-91020-w
11. Cawthorn DM, Steinman HA, Witthuhn RC. DNA barcoding reveals a high incidence of fish species misrepresentation and substitution on the South African market. Food Res Int. 2012;46(1):30-40. https://doi.org/10.1016/j.foodres.2011.11.011
12. Changizi R, Farahmand H, Soltani M, Asareh R, Ghiasvand Z. Species identification reveals mislabeling of important fish products in Iran by DNA barcoding. Iran J Fish Sci. 2013;12(4):783-91. Disponível em:https://aquadocs.org/handle/1834/11666
13. Chen KC, Zakaria D, Altarawneh H, Andrews GN, Ganesan GS, John KM et al. DNA barcoding of fish species reveals low rate of package mislabeling in Qatar. Genome. 2019;62(2):69-76. https://doi.org/10.1139/gen-2018-0101
14. Cutarelli A, Amoroso MG, Roma A, Girardi S, Galiero G, Guarino A et al. Italian market fish species identification and commercial frauds revealing by DNA sequencing. Food Control. 2014;37:46-50. https://doi.org/10.1016/j.foodcont.2013.08.009
15. Deconinck D, Volckaert FAM, Hostens K, Panicz R, Eljasik P, Faria M et al. A high-quality genetic reference database for European commercial fishes reveals substitution fraud of processed Atlantic cod (Gadus morhua) and common sole (Solea solea) at different steps in the Belgian supply chain. Food Chem Toxicol. 2020;141:111417. https://doi.org/10.1016/j.fct.2020.111417
16. Delpiani G, Delpiani SM, Antoni MYD, Ale MC, Fischer L, Lucifora LO et al. Are we sure we eat what we buy? Fish mislabelling in Buenos Aires province, the largest sea food market in Argentina. Fisheries Research. 2020;221:105373. https://doi.org/10.1016/j.fishres.2019.105373
17. Do TD, Choi TJ, Kim J, An HE, Park YJ, Karagozlu MZ et al. Assessment of marine fish mislabeling in South Korea’s markets by DNA barcoding. Food Control. 2019;100:53-7. https://doi.org/10.1016/j.foodcont.2019.01.002
18. Filonzi L, Chiesa S, Vaghi M, Marzano FN. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Research International. 2010;43(5):1383-8. https://doi.org/10.1016/j.foodres.2010.04.016
19. Filonzi L, Vaghi M, Ardenghi A, Rontani PM, Voccia A, Marzano FN. Efficiency of DNA mini-barcoding to assess mislabeling in commercial fish products in Italy: an overview of the last decade. Foods. 2021;10(7):1449. https://doi.org/10.3390/foods10071449
20. Galal-Khallaf A, Ardura A, Mohammed-Geba K, Borrell YJ, Garcia-Vazquez E. DNA barcoding reveals a high level of mislabeling in Egyptian fish fillets. Food Control. 2014;46:441-5. https://doi.org/10.1016/j.foodcont.2014.06.016
21. Gomes G, Correa R, Veneza I, Silva R, Silva D, Miranda J et al. Forensic analysis reveals fraud in fillets from the “Gurijuba” Sciades parkeri (Ariidae – Siluriformes): a vulnerable fish in Brazilian Coastal Amazon. Mitochondrial DNA Part A. 2019;30(5):721-9. https://doi.org/10.1080/24701394.2019.1622694
22. Helyar SJ, Lloyd HD, Bruyn M, Leake J, Bennett N, Carvalho GR. Fish product mislabelling: failings of traceability in the production chain and implications for illegal, unreported and unregulated (IUU) fishing. PLoS One. 2014;9(6):e98691. https://doi.org/10.1371/journal.pone.0098691
23. Kappel K, Schröder U. Substitution of high-priced fish with low-priced species: adulteration of common sole in German restaurants. Food Control. 2016;59:478-86. https://doi.org/10.1016/j.foodcont.2015.06.024
24. Muñoz-Colmenero M, Blanco O, Arias V, Martinez JL, Garcia-Vazquez E. DNA authentication of fish products reveals mislabeling associated with seafood processing. Fisheries. 2016;41(3):128-38. https://doi.org/10.1080/03632415.2015.1132706
25. Sameera S, Jose D, Harikrishnan M, Ramachandran A. Species substitutions revealed through genotyping: implications of traceability limitations and unregulated fishing. Food Control. 2021;123:107779. https://doi.org/10.1016/j.foodcont.2020.107779
26. von der Heyden S, Barendse J, Seebregts AJ, Matthee CA. Misleading the masses: detection of mislabelled and substituted frozen fish products in South Africa. ICES J Mar Sci. 2010;67(1):176-85. https://doi.org/10.1093/icesjms/fsp222
27. Xiong X, Guardone L, Cornax MJ, Tinacci L, Guidi A, Gianfaldoni D et al. DNA barcoding reveals substitution of Sablefish (Anoplopoma fimbria) with Patagonian and Antarctic Toothfish (Dissostichus eleginoides and Dissostichus mawsoni) in online market in China: how mislabeling opens door to IUU fishing. Food Control. 2016;70:380-91. https://doi.org/10.1016/j.foodcont.2016.06.010
28. Yan S, Lai G, Li L, Xiao H, Zhao M, Wang M. DNA barcoding reveals mislabeling of imported fish products in Nansha new port of Guangzhou, Guangdong province, China. Food Chemistry. 2016;202:116-9. https://doi.org/10.1016/j.foodchem.2016.01.133
29. Feitosa LM, Martins APB, Giarrizzo T, Macedo W, Monteiro IL, Gemaque R et al. DNA-based identification reveals illegal trade of threatened shark species in a global elasmobranch conservation hotspot. Sci Rep. 2018;8:3347. https://doi.org/10.1038/s41598-018-21683-5
30. Fox M, Mitchell M, Dean M, Elliott C, Campbell K. The seafood supply chain from a fraudulent perspective. Food Secur. 2018;10(4):939-63. https://doi.org/10.1007/s12571-018-0826-z
31. Calosso MC, Claydon JAB, Mariani S, Cawthorn DM. Global footprint of mislabelled seafood on a small island nation. Biol Conserv. 2020;245:108557. https://doi.org/10.1016/j.biocon.2020.108557
32. Jacquet JL, Pauly D. Trade secrets: renaming and mislabeling of seafood. Mar Policy. 2008;32(3):309-18. https://doi.org/10.1016/j.marpol.2007.06.007
33. Ministério da Agricultura, Pecuária e Abastecimento (BR). Instrução Normativa nº 53, de 1° de Setembro de 2020. Define o nome comum e respectivos nomes científicos para as principais espécies de peixes de interesse comercial destinados ao comércio nacional. Diário Oficial da União. Brasília, DF, 04 set 2020. Seção 1(171):2-5.
34. Goetz FW, Anulacion BF, Arkoosh MR, Cook MA, Dickhoff WW, Dietrich JP et al. Status of sablefish, Anoplopoma fimbria, aquaculture. J World Aquac Soc. 2021;52(3):607-46. https://doi.org/10.1111/jwas.12769
35. Head MA, Keller AA, Bradburn M. Maturity and growth of sablefish, Anoplopoma fimbria, along the U.S. West Coast. Fish Res. 2014;159:56-67. https://doi.org/10.1016/j.fishres.2014.05.007
36. Friesen EN, Balfry SK, Skura BJ, Ikonomou MG, Higgs DA. Evaluation of cold-pressed flaxseed oil as an alternative dietary lipid source for juvenile sablefish (Anoplopoma fimbria). Aquac Res. 2011;44(2):182-99. https://doi.org/10.1111/j.1365-2109.2011.03022.x
37. Vasquez I, Cao T, Hossain A, Valderrama K, Gnanagobal H, Dang M et al. Aeromonas salmonicidainfection kinetics and protective immune response to vaccination in sablefish (Anoplopoma fimbria). Fish Shellfish Immunol. 2020;104:557-66. https://doi.org/10.1016/j.fsi.2020.06.005
38. Rose GA. Reconciling overfishing and climate change with stock dynamics of Atlantic cod (Gadus morhua) over 500 years. Can J Fish Aquat Sci. 2004;61(9):1553-7. https://doi.org/10.1139/f04-173
39. Herrero B, Madriñán M, Vieites JM, Espiñeira M. Authentication of Atlantic cod (Gadus morhua) using real time PCR. J Agric Food Chem. 2010;58(8):4794-9. https://doi.org/10.1021/jf904018h
40. Norberg B, Brown CL, Halldorsson O, Stensland K, Bjornsson BT. Photoperiod regulates the timing of sexual maturation, spawning, sex steroid and thyroid hormone profiles in the Atlantic cod (Gadus morhua). Aquaculture. 2004;229(1-4):451-67. https://doi.org/10.1016/S0044-8486(03)00393-4
41. Tanner SE, Vasconcelos RP, Reis-Santos P, Cabral HN, Thorrold SR. Spatial and ontogenetic variability in the chemical composition of juvenile common sole (Solea solea) otoliths. Estuar Coast Shelf Sci. 2011;91(1):150-7. https://doi.org/10.1016/j.ecss.2010.10.008
42. Parma L, Bonaldo A, Massi P, Yúfera M, Martínez-Rodríguez G, Gatta PP. Different early weaning protocols in common sole (Solea solea L.) larvae: implications on the performances and molecular ontogeny of digestive enzyme precursors. Aquaculture. 2013;414-415:26-35. https://doi.org/10.1016/j.aquaculture.2013.07.043
43. Lacroix G, Maes GE, Bolle LJ, Volckaert FAM. Modelling dispersal dynamics of the early life stages of a marine flatfish (Solea solea L.). J Sea Res. 2013;84:13-25. https://doi.org/10.1016/j.seares.2012.07.010
44. Li W, Chen X, Xu Q, Zhu J, Dai X, Xu L. Genetic population structure of Thunnus albacares in the Central Pacific Ocean based on mtDNA COI gene sequences. Biochem Genet. 2015;53(1-3):8-22. https://doi.org/10.1007/s10528-015-9666-0
45. Anderson G, Lal M, Hampton J, Smith N, Rico C. Close kin proximity in yellowfin tuna (Thunnus albacares) as a driver of population genetic structure in the Tropical Western and Central Pacific Ocean. Front Mar Sci. 2019;6:341. https://doi.org/10.3389/fmars.2019.00341
46. Collette BB, Carpenter KE, Polidoro BA, Juan-Jordá MJ, Boustany A, Die DJ et al. High value and long life: double jeopardy for tunas and billfishes. Science. 2011;333(6040):291-2. https://doi.org/10.1126/science.1208730
47. Al-Hosni AHS, Siddeek SM. Growth and mortality of the narrowbarred Spanish Mackerel, Scomberomorus commerson (Lacepède), in Omani waters. Fish Manag Ecol. 1999;6(2):145-60. https://doi.org/10.1046/j.1365-2400.1999.00134.x
48. Yang R, Han M, Fu Z, Wang Y, Zhao W, Yu G et al. Immune responses of Asian seabass Lates calcariferto dietary Glycyrrhiza uralensis. Animals. 2020;10(9):1629. https://doi.org/10.3390/ani10091629
49. Banerjee I, Sadhu T, Mukherjee R, Bhattacharjee A, Chakrabarty J. Nutritional consequences of sun-drying, freezing, and frying of Lates calcarifer on human health. J Indian Chem Soc. 2021;98(10):100158. https://doi.org/10.1016/j.jics.2021.100158
50. Tan CW, Malcolm TTH, Kuan CH, Thung TY, Chang WS, Loo YY et al. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from short mackerels (Rastrelliger brachysoma) in Malaysia. Front Microbiol. 2017;8:1087. https://doi.org/10.3389/fmicb.2017.01087
51. Senarat S, Kettratad J, Jiraungkoorskul W, Kangwanrangsan N, Amano M, Shimizu A et al. Distribution and changes in the sbGnRH system in Rastrelliger brachysoma males during the breeding season. Sci Mar. 2021;85(3):187-95. https://doi.org/10.3989/scimar.05023.017
52. Ximenes LF. Produção de pescado no Brasil e no nordeste brasileiro. Cad Set ETENE. 2021;5(150). [acesso 2023 Jun 05]. Disponível em: https://www.bnb.gov.br/s482-dspace/bitstream/123456789/649/1/2021_CDS_150.pdf
53. Lopes IG, Oliveira RG, Ramos FM. Perfil do consumo de peixes pela população brasileira. Biota Amazônia. 2016;6(2):62-5. http://dx.doi.org/10.18561/2179-5746/biotaamazonia.v6n2p62-65
54. Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS et al. Mislabelling of a depleted reef fish. Nature. 2004;430(6997):309-10. https://doi.org/10.1038/430309b
55. Gold JR, Voelker G, Renshaw MA. Phylogenetic relationships of tropical western Atlantic snappers in subfamily Lutjaninae (Lutjanidae: Perciformes) inferred from mitochondrial DNA sequences. Biol J Linn Soc Lond. 2011;102(4):915-29. https://doi.org/10.1111/j.1095-8312.2011.01621.x
56. Pinto A, Marchetti P, Mottola A, Bozzo G, Bonerba E, Ceci E et al. Species identification in fish fillet products using DNA barcoding. Fish Res. 2015;170:9-13. https://doi.org/10.1016/j.fishres.2015.05.006
57. Cline E. Marketplace substitution of Atlantic salmon for Pacific salmon in Washington State detected by DNA barcoding. Food Res Int. 2012;45(1):388-93. https://doi.org/10.1016/j.foodres.2011.10.043
58. Xiong X, Yao L, Ying X, Lu L, Guardone L, Armani A et al. Multiple fish species identified from China’s roasted Xue Yu fillet products using DNA and mini-DNA barcoding: implications on human health and marine sustainability. Food Control. 2018;88:123-30. https://doi.org/10.1016/j.foodcont.2017.12.035
59. Miller DD, Clarke M, Mariani S. Mismatch between fish landings and market trends: a western European case study. Fish Res. 2012;121-122:104-14. https://doi.org/10.1016/j.fishres.2012.01.016
60. Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS et al. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314(5800):787-90. https://doi.org/10.1126/science.1132294
61. Cheung WWL, Watson R, Pauly D. Signature of ocean warming in global fisheries catch. Nature. 2013;497(7449):365-8. https://doi.org/10.1038/nature12156
62. Bryndum-Buchholz A, Tittensor DP, Blanchard JL, Cheung WWL, Coll M, Galbraith ED et al. Twenty-first-century climate change impacts on marine animal biomass and ecosystem structure across ocean basins. Glob Chang Biol. 2018;25(2):459-72. https://doi.org/10.1111/gcb.14512
63. Worm B, Hilborn R, Baum JK, Branch TA, Collie JS, Costello C et al. Rebuilding global fisheries. Science. 2009;325(5940):578-85. https://doi.org/10.1126/science.1173146
64. Pauly D, Watson R, Alder J. Global trends in world fisheries: impacts on marine ecosystems and food security. Phil Trans R Soc B. 2005;360(1453):5-12. https://doi.org/10.1098/rstb.2004.1574
65. Martinsohn JT, Raymond P, Knott T, Glover KA, Nielsen EE, Eriksen LB et al. DNA-analysis to monitor fisheries and aquaculture: too costly? Fish and Fisheres. 2019;20(2):391-401. https://doi.org/10.1111/faf.12343
66. Lima FC, Mesquita EFM. Fraudes detectadas na comercialização de pescado no município de Niterói, Estado do Rio de Janeiro, Brasil. Rev Bras Cienc Vet. 1996;3(2):39-43. https://doi.org/10.4322/rbcv.2015.042
67. Anjos NF, Tomita RY. Estudo do valor nutricional do pescado visando agregação de valor e estímulo ao seu consumo. VII Simpósio de Controle de Qualidade do Pescado; outubro de 2016; São Paulo: Universidade Católica de Santos. Disponível em: https://www.researchgate.net/publication/311793865_Estudo_do_Valor_Nutricional_do_Pescado_Visando_Agregacao_de_Valor_e_Estimulo_ao_seu_Consumo
68. Ministério da Agricultura, Pecuária e Abastecimento – MAPA. Manual identifica espécies de peixes para ajudar no combate à fraude. Brasília. 2016 [acesso 2023 Jun 05]. Disponível em: https://www.gov.br/agricultura/pt-br/assuntos/noticias/manual-identifica-especies-de-peixes-para-ajudar-no-combate-a-fraude
69. Focardi S. Levels of mercury and polychlorobiphenyls in commercial food in Siena Province (Tuscany, Italy) in the period 2001-2010. Microchem J. 2012;105:60-4. https://doi.org/10.1016/j.microc.2012.01.013
70. Giusti A, Castigliego L, Rubino R, Gianfaldoni D, Guidi A, Armani A. A conventional multiplex PCR assay for the detection of toxic gemfish species (Ruvettus pretiosus and Lepidocybium flavobrunneum): a simple method to combat health frauds. J Agric Food Chem. 2016;64(4):960-8. https://doi.org/10.1021/acs.jafc.5b04899
71. Cohen NJ, Deeds JR, Wong ES, Hanner RH, Yancy HF, White KD et al. Public health response to puffer fish (tetrodotoxin) poisoning from mislabeled product. J Food Prot. 2009;72(4):810-7. https://doi.org/10.4315/0362-028X-72.4.810
72. Garcia-Vazquez E, Machado-Schiaffino G, Campo D, Juanes F. Species misidentification in mixed hake fisheries may lead to overexploitation and population bottlenecks. Fish Res. 2012;114:52-5. https://doi.org/10.1016/j.fishres.2011.05.012
73. Nedunoori A, Turanov SV, Kartavtev YP. Fish product mislabeling identified in the Russian far east using DNA barcoding. Gene Rep. 2017;8:144-9. https://doi.org/10.1016/j.genrep.2017.07.006
74. Brito MA, Schneider H, Sampaio I, Santos S. DNA barcoding reveals high substitution rate and mislabeling in croaker fillets (Sciaenidae) marketed in Brazil: the case of “pescada branca” (Cynoscion leiarchus and Plagioscion squamosissimus). Food Res Int. 2015;70:40-6. https://doi.org/10.1016/j.foodres.2015.01.031
75. Veneza I, Silva R, Freitas L, Silva S, Martins K, Sampaio I et al. Molecular authentication of Pargo fillets Lutjanus purpureus (Perciformes: Lutjanidae) by DNA barcoding reveals commercial fraud. Neotrop Ichthyol. 2018;16(1):e170068. https://doi.org/10.1590/1982-0224-20170068
76. Staffen CF, Staffen MD, Becker ML, Lofgren SE, Muniz YCN, Freitas RHA et al. DNA barcoding reveals the mislabeling of fish in a popular tourist destination in Brazil. Peer J. 2017;5:e4006. https://doi.org/10.7717/peerj.4006
77. Galimberti A, Mattia F, Losa A, Bruni I, Federici S, Casiraghi M et al. DNA barcoding as a new tool for food traceability. Food Res Int. 2013;50(1):55-63. https://doi.org/10.1016/j.foodres.2012.09.036
78. Ministério da Agricultura, Pecuária e Abastecimento – MAPA. Manual de inspeção para identificação de espécies de peixes e valores indicativos de substituições em produtos da pesca e aquicultura. Brasília. 2022 [acesso 2023 Jun 05]. Disponível em: https://wikisda.agricultura.gov.br/pt-br/Inspe%C3%A7%C3%A3o-Animal/Manual-de-procedimentos-de-inspecao-e-fiscalizacao-de-pescado-e-derivados-em-estabelecimentos-sob-inspecao-federal
79. Carvalho DC, Guedes D, Trindade MG, Coelho RMS, Araujo PHL. Nationwide Brazilian governmental forensic programme reveals seafood mislabelling trends and rates using DNA barcoding. Fish Res. 2017;191:30-5. https://doi.org/10.1016/j.fishres.2017.02.021
80. Ministério da Agricultura e Pecuária – MAPA. Operação Semana Santa fiscaliza qualidade do pescado em 23 estados e no DF. Brasília. 2023 [acesso 2023 Jun 05]. Disponível em: https://www.gov.br/agricultura/pt-br/assuntos/noticias/operacao-semana-santa-fiscaliza-qualidade-do-pescado-em-23-estados-e-no-df

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Derechos de autor 1969 Carolina Laipelt Matias, Andrea Troller Pinto, Juliana Querino Goulart